MAGNET Documentation
Usage Instructions
Step 1: Specify how many query gene lists/clusters you are submitting

Step 2: Upload your query gene lists/clusters
Input the gene sets through either text area or file upload. If you selected multiple clusters in Step 1, only file upload will be available.
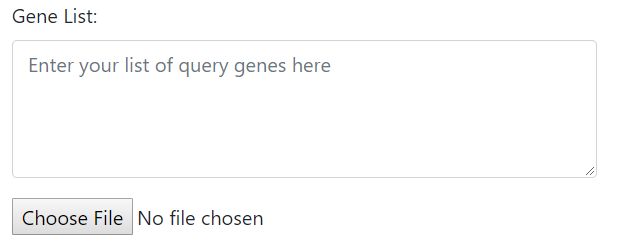
Acceptable input files are csv files (other allowed file formats will be implemented) with gene names in the 1st column, which can be either MGI gene symbols or ENSEMBL IDs, and user-assigned set/cluster number in the 2nd column. The 2nd column is not required if you are submitting only one gene set.

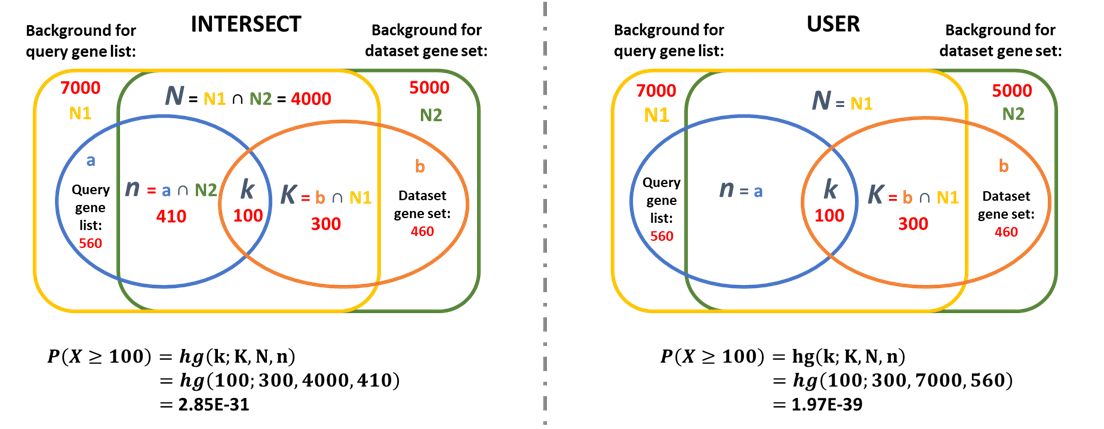
Step 3: Choose background calculation mode

- If INTERSECT is selected, the overlap between user supplied background and the genes associated in each datasets will be used as background
- If USER is selected, the user supplied background gene list will be used as background
More description on the statistical tests being performed can be found in the Explanation of Output tab.
Step 4: Upload your background gene list
Paste here a list of background genes, in either MGI symbols or ENSEMBL IDs.

If using file upload, input the gene names in the 1st column of the csv file.
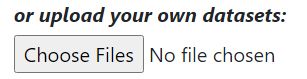
Step 5: Choose datasets you desire to be analyzed by MAGNET
Simply deselect any dataset you don't want to be processed by MAGNET. Click the names to gain more info on each dataset.

Alternatively, submit custom datasets you wish to test agasint by file upload. The file format requirements are the same as in step 2.
Step 6: Press submit button

Frequently Asked Questions
Question: What does MAGNET stands for?
Answer: Macrophage Annotation of Gene Network Enrichment Tool
Question: Do MAGNET support human genome?
Answer: MAGNET only supports mouse genome at the moment. However, we plan to implment this feature in the future.
Question: How often will MAGNET be updated?
Answer: There are no fixed schedules, but MAGNET database will be updated regularly as relavent macrophage related manuscripts and improved gene annotations become available.
Question: What gene name formats do MAGNET support?
Answer: MAGNET supports ENSEMBL gene IDs and MGI gene symbols.
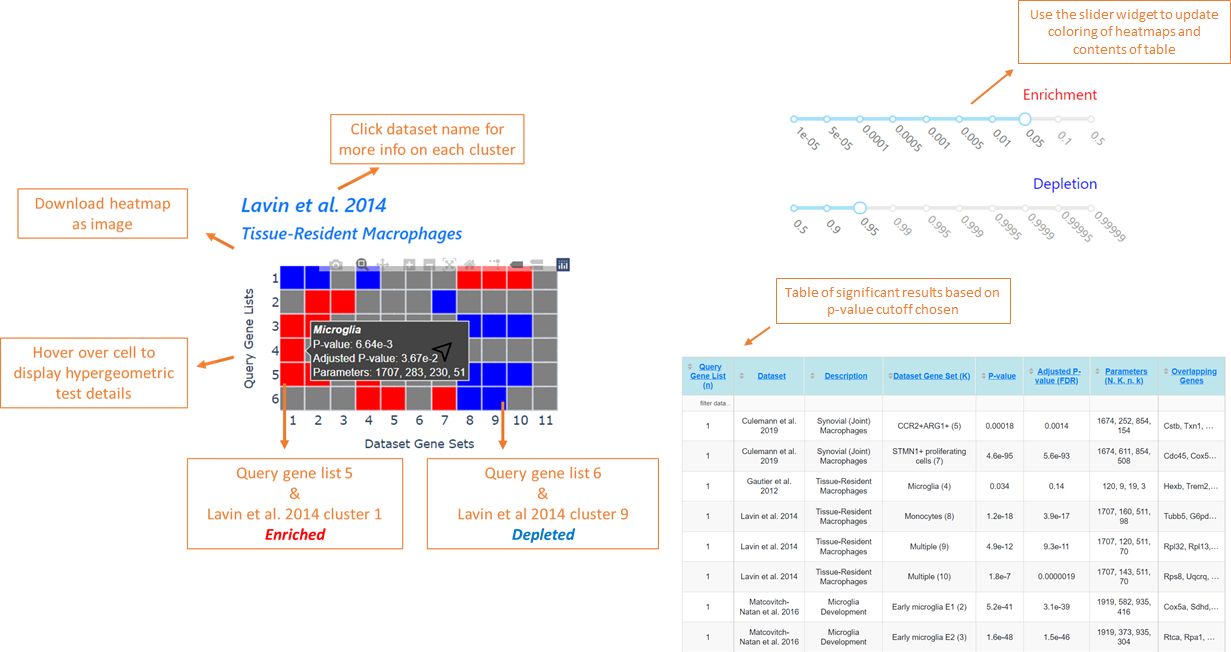
Explanation of Output
Interpreting the results from MAGNET
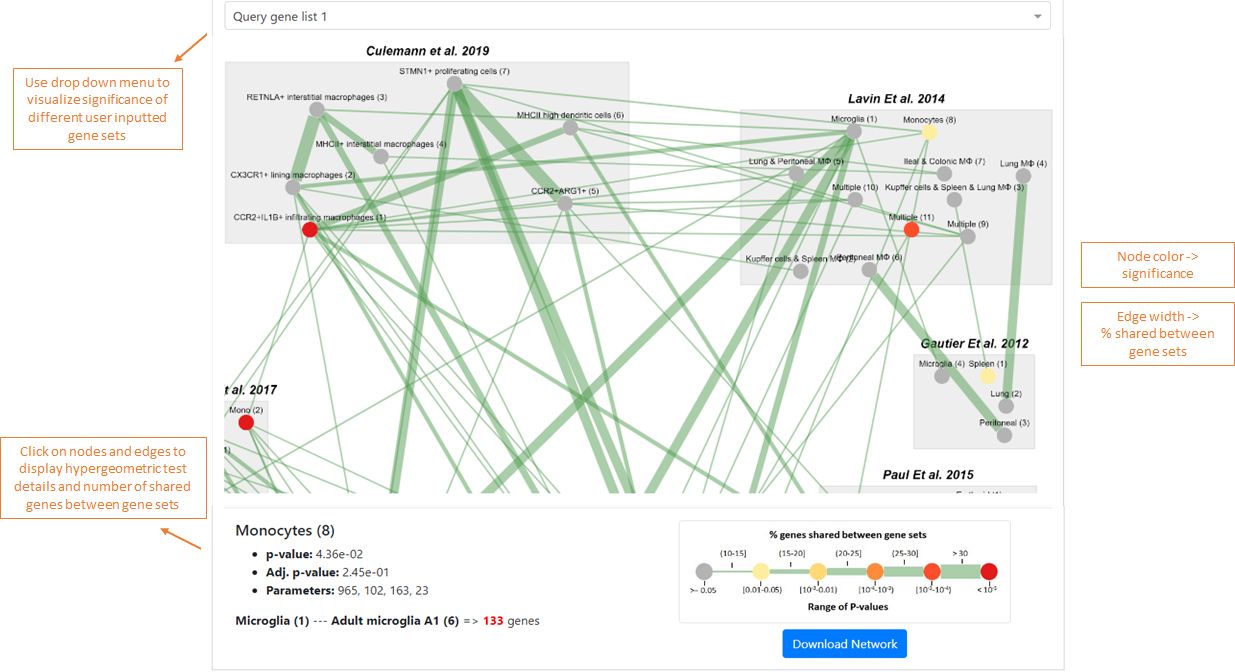
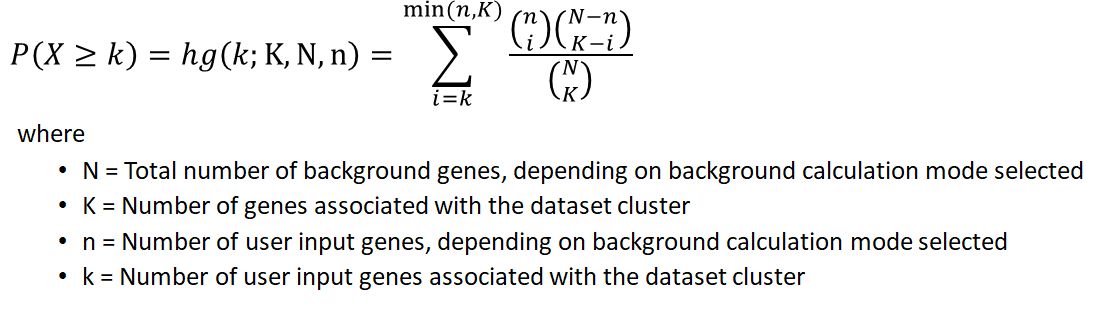
Intro to the enrichment test implemented in MAGNET
Similar to Gene Ontology (GO) analyses, MAGNET utilizes the hypergeometric distribution to assess significance, given by the following formula:
Let's say an user obtained a list of differentially regulated genes in macrophages during infection (DE), and want to find out if they are enriched for inflammatory genes annotated in an unrelated dataset (IF).

Depending on what background calculation mode is selected, their respective venn diagram illustrations and hypergeometric p-values will be: